
EQUATIONS TO BE SOLVED AND BOUNDARY CONDITIONS
Main Plasma
Before starting the run you need to select the type and value of the boundary
conditions for all equations. Please note that the value should correspond to
the type. All equations allow for following types of boundary conditions:
- OFF - equation is not solved, initial profiles will be kept for whole run
- value - edge value should be specified
- gradient - edge gradient should be specified
- scale_length - edge scale length should be specified
- generic - generic form: a1*y´ + a2*y = a3 of the boundary condition is assumed, 3 coefficients (a1,a2,a3) should be provided
- value_from_input_CPO - equation is solved, edge value evolution will be red from input shot
- profile_from_input_CPO - equation is not solved, profile evolution will be red from input shot
The particular equation will be activated if the boundary condition type for
it is other than OFF

To set up boundary conditions:
- right click on the box BEFORE THE TIME EVOLUTION
- select Configure actor
- select appropriate boundary condition for each equation
- specify values for boundary conditions corresponding to the type and to the ion component
- Commit
The workflow will not allow the user all particle components (ions[1:NION]+electrons) to be run predictively. At least one of them shall be set to OFF (this component will be computed from quasi-neutrality condition).
!!! If electron density is solved, all ions with ni_bnd_type=OFF will be computed from the quasineutrality condition and scaled proportional to specified ni_bnd_value or inversely proportional to their charge, charge_proportional. This is defined by option: ni_from_quasineutrality.
Impurity
You can set up the boundary conditions for impurity ions in a similar way as
for main ions.
!!! Note, that at the moment only types: OFF; value and value_from_input_CPO are accepter by impurity
solver.
To set up boundary conditions:
- right click on the box BEFORE THE TIME EVOLUTION
- select Configure actor
- select appropriate boundary condition for each impurity species (OFF-equation is not solved)
- specify values for boundary density of each impurity component [1:MAX_Z_IMP], separated by commas
- Commit
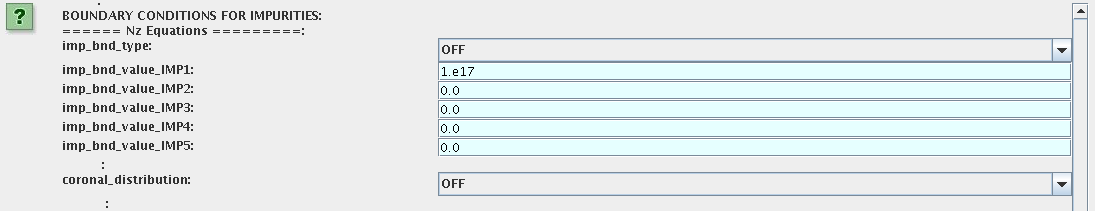
Interface for impurity boundary condition has additional option, coronal_distribution, that allow to preset the edge
values or entire profiles of individual ionization states from coronal distribution. In tis case only single value is required to be specified for
each impurity boundary value.
The options are:
- OFF - the boundary values for impurity densities will be as they are specified above;
- boundary_conditions - the boundary densities will be renormalized with corona, using the first element from above as a total density
- boundary_conditions_and_profiles - the boundary densities and starting profiles will be renormalized with corona, using the first element from above as a total density
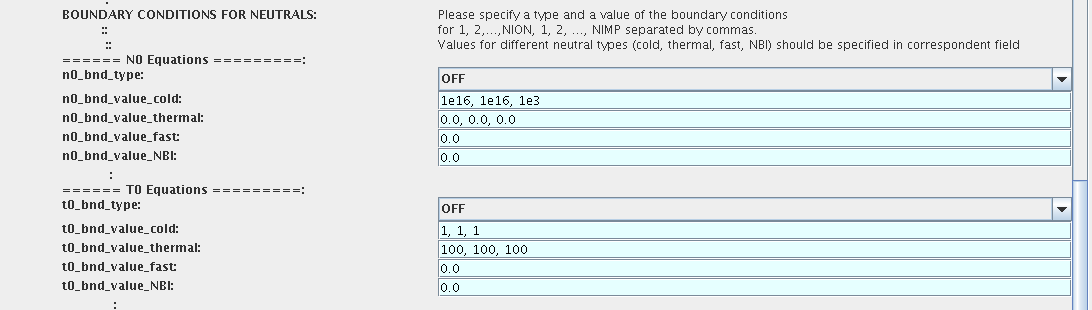
Neutrals
!!! AT THE MOMENT BOUNDARY CONDITIONS FOR NEUTRAL VELOCITIES ARE DISABLED, MIGHT BE ADDED ON REQUEST
Note, that ALL values should be specified in the order: {1, 2, 3 ...NION, 1, 2, 3, ...NIMP}
To set up boundary conditions:
- right click on the box BEFORE THE TIME EVOLUTION
- select Configure actor
- select appropriate boundary condition for each neutral species (OFF-equation is not solved)
- specify values for boundary density and temperature of each neutral component [1, 2, 3 ...NION, 1, 2, 3, ...NIMP], separated by commas
- Commit
Input profiles interpolation
You are going to start the ETS run from some input shot, which might contain
some conflicting rho grids saved to different CPOs.
Thus there is a choice for the user to decide on the grid on which the
starting profiles should be load by the
worflow, Interpolation_of_input_profiles.
To define the interpolation grid select:
- on_RHO_TOR_grid - interpolate input profiles based on the grid specyfied in [m];
- on_RHO_TOR_NORM_grid - interpolate input profiles based on normalised rho grid [0:1]
